Potential growth of the community
The potential growth of the plant community
In the last equation the values are converted to the dimensionless leaf area index by multiplying the term with 0.1.
maximum radiation use efficiency [kg MJ⁻¹] light extinction coefficient [-] is the reduction factor of if the community weighted mean height equals 0.2 m [-]
Visualization
- Combined effects of the total leaf area index on the fraction of photosynthetically active radiation intercepted by plants and how this fraction is reduced when the community height is low due to stronger shading effects.
| extinction coefficient γ_RUE_k | 0.6 | |
| effect of community height on shading α_RUE_cwmH | 0.75 | |
| community weighted mean height H_cwm | 0.7 |
- Influence of specific leaf area and aboveground biomass proportion on the leaf area index, all species have a total biomass of 2000 [kg ha⁻¹] and the aboveground biomass is assumed to be total biomass times aboveground biomass proportion:
. Note, that during the simulation the aboveground biomass proportion is often lower than the trait .
show code
julia
import GrasslandTraitSim as sim
using CairoMakie
using Unitful
let
traits = sim.input_traits()
nspecies = length(traits.sla)
LAIs = zeros(nspecies)
biomass = fill(2000.0u"kg/ha", nspecies)
above_biomass = traits.abp .* biomass
for s in eachindex(LAIs)
LAIs[s] = uconvert(NoUnits, traits.sla[s] * above_biomass[s] * traits.lbp[s] / traits.abp[s])
end
idx = sortperm(traits.sla)
LAIs_sorted = LAIs[idx]
sla = ustrip.(traits.sla[idx])
abp = (traits.abp)[idx]
colorrange = (minimum(abp), maximum(abp))
colormap = :viridis
fig = Figure()
ax = Axis(fig[1, 1]; xlabel = "Specific leaf area [m² g⁻¹]", ylabel = "Leaf area index [-]", title = "")
sc = scatter!(sla, LAIs_sorted, color = abp, colormap = colormap)
Colorbar(fig[1,2], sc; label = "Aboveground biomass per total biomass [-]")
end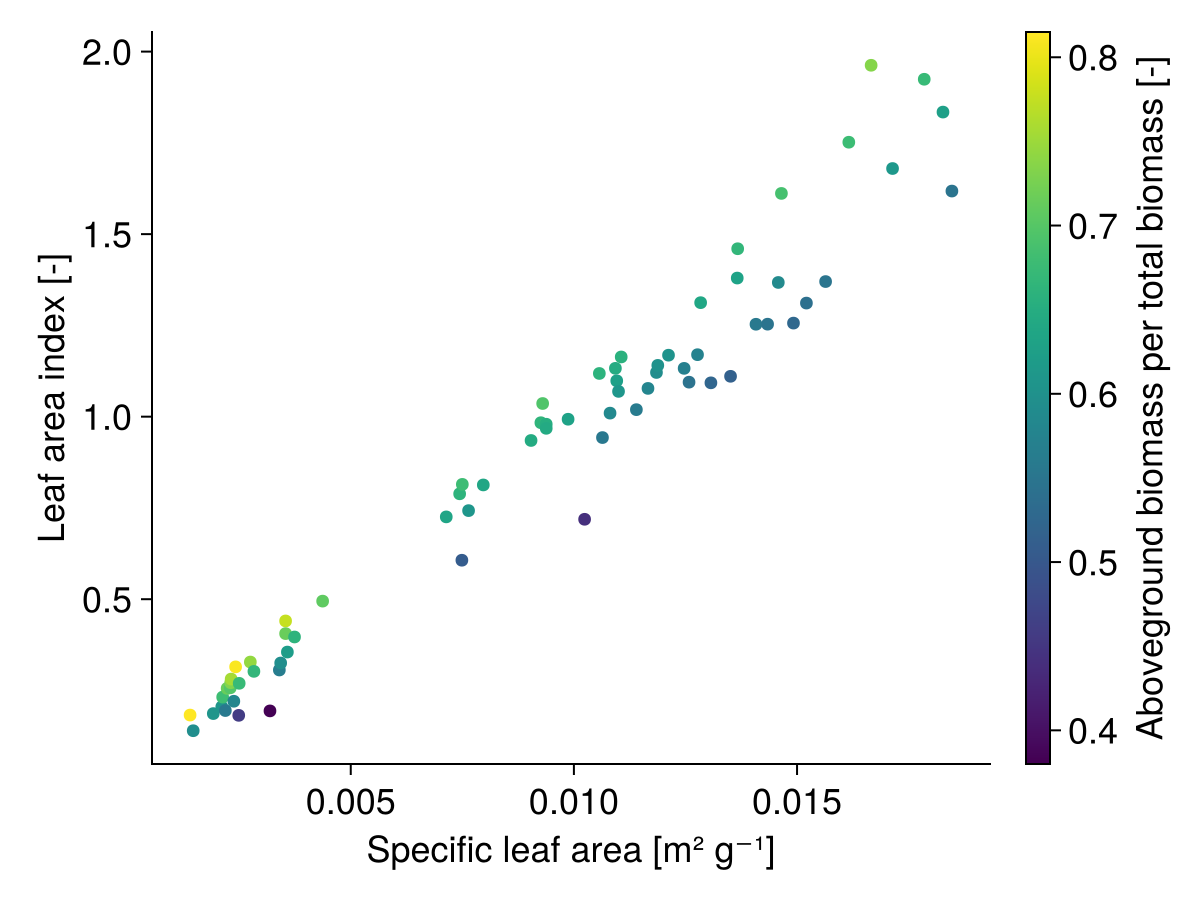
API
GrasslandTraitSim.potential_growth! Function
julia
potential_growth!(
;
container,
above_biomass,
actual_height,
PAR
)Calculate the total potential growth of the plant community.
GrasslandTraitSim.calculate_LAI! Function
julia
calculate_LAI!(; container, above_biomass)Calculate the leaf area index of all species.